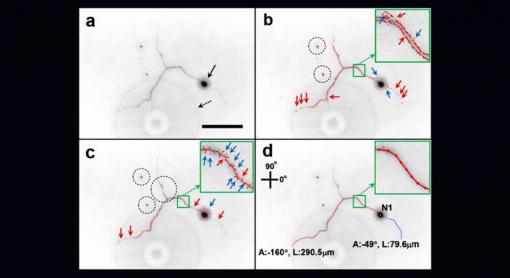
Delicate neuron structures can be hard to spot in microscope photos. The Neuron Image Analyzer (d) does a better job of picking out those structures compared to existing algorithms (b and c). Box a is a raw neuron image.
Credit: Palmore Lab/Brown University
PROVIDENCE, R.I. [Brown University] — Scientists looking for ways to stimulate the growth of neurons can spend hours painstakingly analyzing microscope images of cells growing in petri dishes. A new algorithm developed by Brown University researchers automates that process and analyzes images more accurately than previous automated approaches.
The algorithm and an initial round of testing are described in Nature Scientific Reports.
As neurons grow, they extend stringy appendages called neurites that form critical connections with neighboring cells. These networks of neurons and neurites are essential for healthy nervous system function, and scientists are interested in finding new ways to encourage neuron growth through drugs, electrical stimulation, or other means. To test the effects of those efforts, scientists grow neurons in the lab and apply different treatments to see if they stimulate growth. That usually involves taking hundreds of microscope pictures of neurons as they grow over the course of hours or days.
"You're left with this gigantic stack of photos," said Tayhas Palmore, professor of engineering at Brown and the new paper's senior author. "You need to analyze the changes from one image to the next, and that can be really arduous."
The details in those images are critical. Neurites are tiny structures that are hard to see under a microscope during live-cell imaging. But accurately measuring their length and thickness is important in assessing stimulated cell growth. There are a few algorithms available that automate the image analysis, but they don't do a terribly good job. They generally work by looking at individual pixels in an image and applying a uniform filter that picks out pixels with the highest intensity. Those high-intensity pixels are assumed to be neuron and neurite structures.
The problem is microscope images often are not high quality, making it hard to discern cell structures from random artifacts that may be present in the image. As a result, the filters often include pixels that aren't relevant to neuron structures and edit out pixels that are important. This is especially a problem in measuring tiny neurite appendages. The filters often fail to measure the full extent of neurite growth.
Kwang-Min Kim, a former graduate student in Palmore's lab and now a postdoctoral researcher at Stanford, wanted to find a better solution. Inspired by the previous work of Kilho Son, a graduate student in computer vision and co-first author on the paper, Kim developed a new method that dispenses with the uniform filters used in other approaches. Instead, the new approach, called Neuron Image Analyzer (NIA), takes into account how pixels are related to neighboring pixels.
"We don't just look for high-intensity pixels," Kim said. "We look at the relational information between pixels. This way we can trace pixels that are connected to each other, which helps us trace the entire neuron structure."
Another technique employed by the algorithm uses a particular statistical test that is good at picking out circular or elliptical structures. That test is used to accurately locate and measure the soma, the blob-shaped main body of a neuron.
The researchers tested NIA against existing algorithms, using hand annotation of images as a benchmark. The results showed NIA to be 80 percent as accurate as hand coding, while the other algorithms were only 50 to 60 percent as accurate.
The team hopes that other researchers will make use of the new approach. It could be especially useful in labs that lack the sophisticated and expensive equipment to take extremely high-quality neuron images.
"We want to make this approach available to anyone who is interested in analyzing neuron images, regardless of the quality of their images," Kim said.
Kim and Son plan to continue developing NIA in the hope of further improving its accuracy and speed.
- by Kevin Stacey